General concerns
Q: How to cite the server?
- For
publications, please use the following papers
| Server used |
Citation |
| POLYVIEW-2D |
Porollo A, Adamczak R, Meller J:
POLYVIEW: A Flexible Visualization Tool for Structural and Functional Annotations of Proteins,
Bioinformatics 2004, 20: 2460-2462.
|
| POLYVIEW-3D |
Porollo A, Meller J:
Versatile Annotation and Publication Quality Visualization of Protein Complexes Using POLYVIEW-3D,
BMC Bioinformatics 2007, 8: 316.
|
| POLYVIEW-MM |
Porollo A, Meller J:
POLYVIEW-MM: web-based platform for animation and analysis of molecular simulations,
Nucleic Acids Research 2010, 38: W662-W666.
|
| CoeViz |
Baker FN, Porollo A:
CoeViz: a web-based tool for coevolution analysis of protein residues,
BMC Bioinformatics 2016, 17: 119.
|
- For
electronic resources, please use the corresponding hyperlinks
| Server used |
URL |
| POLYVIEW-2D |
https://polyview.cchmc.org/ |
| POLYVIEW-3D |
https://polyview.cchmc.org/polyview3d.html |
| POLYVIEW-MM |
https://polyview.cchmc.org/conform.html |
| CoeViz |
https://polyview.cchmc.org/ |
- When using the images generated by POLYVIEW-3D, it is highly
recommended to cite also the respective program (PyMol or
RasMol) used for the rendering of a protein structure.
Q: How to cross-link the server?
- First of all, never bookmark or post at your web-site a hyperlink to
the resulting page with the image, as POLYVIEW-2D does
not contain necessary information in the address line of
the browser to retrieve the same page, whereas
POLYVIEW-3D automatically deletes generated files after
two weeks.
- For simple link to the submission form with PDB ID as a
parameter, include the following hyperlink into your
electronic document:
<a href="https://polyview.cchmc.org?PDBName=XXXX">POLYVIEW</a>
or
<a href="https://polyview.cchmc.org/polyview3d.html?PDBName=XXXX">POLYVIEW-3D</a>
for POLYVIEW and POLYVIEW-3D, respectively. XXXX is a PDB ID
represented by a 4-letter code. Example of this
cross-link can be found in the
PDBwiki pages.
- For advanced link with specific requirements, especially when
requesting the visualization of custom coordinate files
in the PDB format,
contact
the developers of the server
in order to set up an automated way for data exchange.
Example of this cross-link can be found in the
MolMovDB
resulting pages.
- To have POLYVIEW handy on your own web-page, you can use the
following code to include the POLYVIEW widget:
<script type="text/javascript">
<!-- width of the widget (px or %) -->
var pw_width = '200px';
<!-- background color of widget header -->
var pw_color_header = '#404040';
<!-- background color of widget body -->
var pw_color_body = '#808080';
<!-- delay between examples (seconds), 0=hide examples, -1=no examples -->
var pw_time_period = 15;
</script>
<script type="text/javascript" src="https://polyview.cchmc.org/pwidget.js"></script>
Here is how it looks with default parameters.
Q: Is stand-alone version of POLYVIEW-3D available?
POLYVIEW-3D is a set of perl scripts combining the rendering programs
with the protein structure analysis tools and databases. In
order to work locally, please use the respective software and
databases freely available for download from their home sites.
Q: How is the privacy of requests and results maintained?
- For the servers usage statistics, we keep track of the user`s ip address,
type of request and time of submission. These data are
logged in addition to the standard access log generated
by Apache.
- When submitted a protein not deposited to PDB, its structure is
kept at our local drive for two weeks only. It is used to
generate images and obtain structural annotations during this
period without asking the user to re-submit the structure upon
each next request.
- All temporary files including submitted protein structures,
calculations, images, and other results to be retrieved to user
are stored in the
directories directly inaccessible from internet and can only be
retrieved using CGI-scripts by following the links provided
personally to user. File names comprise of two parts and formed
by using the time function combined with random number generator
yielding unique and big numbers of different digit length. Thus,
files can not be scanned by internet crawlers, neither file
names can be guessed by the
simple number enumeration. After two weeks since initial request,
all related files are automatically deleted from the disk.
- E-mail addresses, if optionally specified for the POLYVIEW-3D
requests, are used solely for sending the links to user in order
to retrieve results when they are ready.
- Developers of the POLYVIEW server reserve the right to periodically look
through generated results and images randomly chosen among
available in order to check the proper server functioning and
to detect the possible bugs in the server-related software.
Q: Why is the status of my request shown as "waiting in the queue" for long time?
All our servers run on the cluster of more than 100 compute
nodes. However, as these services are free, they do not
have any separate queue or high priority while
sharing computational resources with local
users. Occasionally, cluster is full of requests from
both local and internet users. Just be patient, or specify
your email address to receive a notice once your request
has been processed.
Setting the parameters
Q: PyMol (or RasMol) provides the option that I
can't find at the POLYVIEW-3D request form. What should
I do?
POLYVIEW-3D never was conceived as a complete web-interface to any of
the protein structure rendering programs used in
it. Instead, it is focused on combining the easy access to
high quality and animated images with the automated protein
structure and function annotations. For advanced
rendering and more custom images, please consider using
the stand-alone versions of respective rendering
software. However, scripts provided along with images
generated by POLYVIEW-3D can serve as a starting point
in practical learning the command syntax of PyMol or RasMol.
Q: How can I get bigger images than those available from options?
We restricted the maximal image size to 1000x1000 pixels in order to
avoid creating the huge files when animations requested. However,
one still can get bigger static images (2000x2000 pixels) by
requesting the animations and checking the
HD slides checkbox at the
Animation Settings field set.
Dealing with output results
Q: When I request an animation from POLYVIEW-3D, I see some static slides of the fixed size below the animation. Why?
Along with animated image of requested size, we provide the user with
all slides of maximal size (1000x1000 pixels) that were used
during the preparation of animation. So one can download the
high-quality static image of the desired protein structure
projection in PNG or TIFF format, with 300 DPI resolution in
latter case, to be used for publication.
Q: Why is the image no longer animated, when I copy and paste an animation from POLYVIEW-3D to PowerPoint?
When in the slide designing mode, PowerPoint shows all animations as
static images. Just start your presentation, and the
corresponding images will be shown animated.
Q: I submitted a request to POLYVIEW-3D but obtained a blank image. What is wrong?
- First, make sure that you requested at least one protein chain to be
rendered, as some combinations of options can make all chains
become hidden.
- Second and most typical case, the requested protein is represented by low
resolution model or crystal structure, i.e.
alpha-carbons are available only. Default rendering option,
Cartoon, fails to generate any image of such a
structure. Try to change the chain rendering to the
CPK spacefill or Surface options.
See example below.
|
Changing the rendering options for low resolution protein structure
(PDB code 2lzh)
|
Rendering with
default Cartoon option

|
Rendering with
CPK spacefill option

|
Rendering with
Surface option

|
Q: I requested a protein structure to be rendered
with transparent surface. Why did the color of the chain change
to the darker tint?
It is a part of PyMol rendering. When transparency requested along
with black or, in general, dark background, all colors used
for surface renderings become shaded. To
avoid this effect, change the background of the image to white
or other light color. See example below.
|
Changing the image background to keep the original colors of
protein structure rendered with transparent elements
(PDB code 1mba)
|
Rendering with default Black background
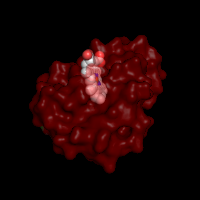
|
Rendering with White background
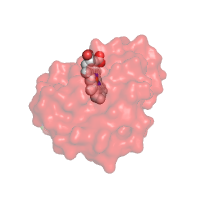
|




