POLYVIEW-3D Tutorial
Part 1. Source of structural data
We start by reviewing the set of options that allows one to specify a PDB formatted file with the macromolecule of interest (e.g., protein or DNA/RNA) to be used as input for rendering.
Data source
Both the Protein Data Bank
(PDB)
deposited entries and the
PDB formatted
custom files can be submitted to the POLYVIEW
server. Lines with ATOM cards are the only
required data to be present in the file. For registered
PDB entries, it is sufficient to enter a 4-letter code
(e.g., 1a4y). NOTE: when both a PDB code
and a custom PDB file are specified, the server will use
only the PDB code as input, and will ignore the submitted file.
For protein structures registered in PDB, one can choose between the
asymmetric unit and the
biological unit if the
latter is available from the
(PISA)
server. Below is example of difference between
asymmetric and
biological units, using
the structure of a porin from Rh. blastica
(PDB id 1bh3).
NOTE: Definition of biological unit is not a straight forward task and may not be always defined unambiguously. That is why different sources may sometimes provide alternative multimeric structures. The user can refer to Proteopedia to read more on this issue. Furthermore, if someone is not satisfied with definition of biological unit by PISA, one can create an alternative model, e.g., using the MakeMultimer utility, and subsequently submit it as a custom file to POLYVIEW-3D for visualization.
| Asymmetric unit (Default option) |
Biological unit |
|---|---|
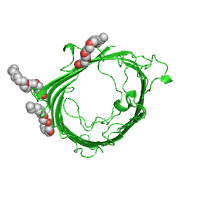
|
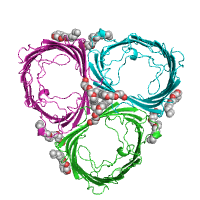
|
| Click on respective image to see options used for its rendering. | |
Last modified: Thu Feb 9 13:22:22 EST 2012