POLYVIEW-3D Tutorial
Part 3. Chain rendering settings
Using the set of options described in this section, one can define how each chain is to be represented. Chains can be specified individually or in groups. Several options can be set for the same chain, if combined rendering is needed. For example, sticks can be overlaid with the cartoon representation.
Individual chains settings
For each chain of a macromolecular structure it is possible to specify its rendering style and coloring scheme. In particular, a protein chain can be shown as:
-
Cartoonrepresenting secondary structure elements formed by a chain backbone. -
Wireframewhen all bonds including side chains are represented by sticks. -
CPK spacefillrepresenting atoms as spheres of different radii depending on an atom type. -
Balls and sticksshowing atoms and chemical bonds as spheres and sticks, respectively. -
Smooth
surface, which is available with the PyMol rendering program only, whereas the use of RasMol with this setting will make chain rendered in spherical representation (CPK). -
And finally, a chain can be set to be
hidden, for example if it is not of interest in context of the image.
To specify a chain to be customized, one needs to enter its label
(single character) in the corresponding text field. In the
case of unlabelled chains, a space or dash (minus) sign
should be used. Row(s) with per-chain settings will be
ignored by the server if no chain label is specified.
NOTE: use the Chains Lookup
button to see available chain labels for a given input.
A number of coloring schemes are available for showing chains under different perspectives. They include simple profiles, such as hydrophobicity or temperature factors, and more complex annotations that involve additional analysis, e.g. evolutionary conservation profiles or mapping interacting residues from homologous proteins to the query chain.
Currently, the following coloring schemes are available:
-
By custom color− allows one to pick a color from the color grid, and to apply it to the corresponding chain. In addition, the selected color is also shown in HTML format for reference. -
By atom type− can be used to color the atoms of the backbone and side chains according to their properties. -
By B-factor− shows temperature factors optionally included in PDB files to estimate the extent of thermal fluctuations and identify rigid, as well as relatively flexible regions of the structure. Note that each rendering program has its own coloring scheme for representing temperature factors. Moreover, many programs or web-servers use B-factor field to encode some other information, e.g., conservation scores or class assignments. POLYVIEW-3D accepts prediction results in such a way from two web-servers, SPPIDER and ConSurf (see legends below). -
By conservation− invokes a PsiBLAST run to derive multiple sequence alignment against nr database and to calculate the corresponding position specific conservation scores, as defined in the Residue highlighting settings section. All residues within the chain are colored according to their conservation, see the color scale below. -
By interaction− performs an automatic search for close sequence homologs of the query chain deposited in PDB, and involved in different types of interactions. Interacting residues with interchain contacts are identified and mapped to the corresponding residues of the query chain. Interacting sites are colored according to the type of interaction (see legend below), whereas the rest of the chain is white. Note, that due to mapping from multiple complexes, the same residue may be found to interact with another protein chain or a DNA molecule, depending on the complex. -
By hydrophilicity− colors residues by the hydrophobicity, polarity, charge and combination of these properties, see amino acid assignments below. -
By acidity− discriminates basic and acidic residues, other amino acids are colored in white, see assignments below. -
By spectrum− renders a chain using rainbow colors with blue for N-terminus and red for C-terminus. -
By secondary structure− colors amino acid residues according to protein secondary structure they are in a given protein as defined using the DSSP program. Helices are colored red, beta-sheets are green, and coils are blue. Note that secondary structures defined by DSSP may differ from definitions and thus structure representations by rendering programs (PyMol and RasMol).
To add or delete rows of per-chain settings, use the corresponding
+ and
− buttons on the
right hand side of the option text fields.
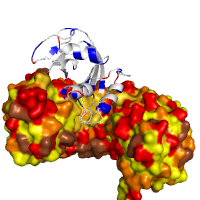
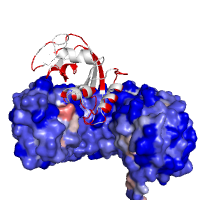
Images below demonstrate several alternative ways of rendering protein chains along with different color schemes applied. As an example, a high affinity protein complex between human placental RNase inhibitor (hRI) and vessel-inducing human angiogenin (Ang) is shown under various perspectives (PDB id 1a4y). Panel in the middle might help explaining the high binding affinity due to extended electrostatic interactions.
Other chains settings
All chains not specified in individual
chain settings fall into the group called
rest chains. And the same
rendering settings, as well as coloring schemes, defined in
previous section can be applied to them as a group with a
few differences. Analog of
By custom color option is
called All in same color
and allows one to shade the remaining chains down from
those of special interest using the same color. In the
case of structures with multiple chains, it is convenient
to select coloring scheme
By chain that makes
rendering programs automatically assign different colors
to each chain. NOTE:
PyMol
and
RasMol
apply their own conventions on colors when performing this
function.
Caution should be exerted when applying coloring schemes
By conservation and
By interaction
to the group
rest chains. In the case
of multichain structures, it may require a considerably
long period of time to fully process this kind of request.
Last modified: Thu Feb 9 14:02:01 EST 2012